Jin XU
Biography
I did my postdoc training from 2019 to 2024 at the University of Washington in Seattle, USA.
I did my Ph.D. study at Pohang University of Science and Technology (POSTECH) and Aisa Pacific Center for Theoretical Physics (APCTP) in Pohang, Korea.
I did my Bachelor's and Master's study at Harbin Institute of Technology in Harbin, China.
Research Interests
My research interests focus on data science, software development, and computational modeling in complex systems or networks, with applications in biomedicine. Examples of the past research in biomedicine include the pancreatic islet system for diabetes, the immune system for infectious disease, and signaling networks for cancer.
Data science
Software development
Computational modeling
Publications (Selected)
Journal Articles:
 |
Xu, J.*, Smith, L..
Curating Models from BioModels: Developing a Workflow for Creating OMEX Files..
In: PLOS ONE, 2024.
(JCR Q1, Impact Factor: 2.9)
[Paper] [Code]
|
 |
Xu, J.*, Wiley, H. S., Sauro, H. M..
Generating Synthetic Signaling Networks for in Silico Modeling Studies.
In: Journal of Theoretical Biology, 2024.
(JCR Q2, Impact Factor: 1.9)
[Paper] [Preprint] [Code]
|
 |
Xu, J.#, Geng, G.#, et. al..
SBcoyote: An Extensible Python-Based Reaction Editor and Viewer.
In: Biosystems, 2023.
(JCR Q2, Impact Factor: 2.0) [# Equal contribution]
[Paper][Preprint] [Code]
|
 |
Xu, J.*.
SBMLKinetics: A Tool for Annotation-Independent Classification of Reaction Kinetics for SBML Models.
In: BMC Bioinformatics, 2023.
(JCR Q1, Impact Factor: 2.9)
[Paper] [Code]
|
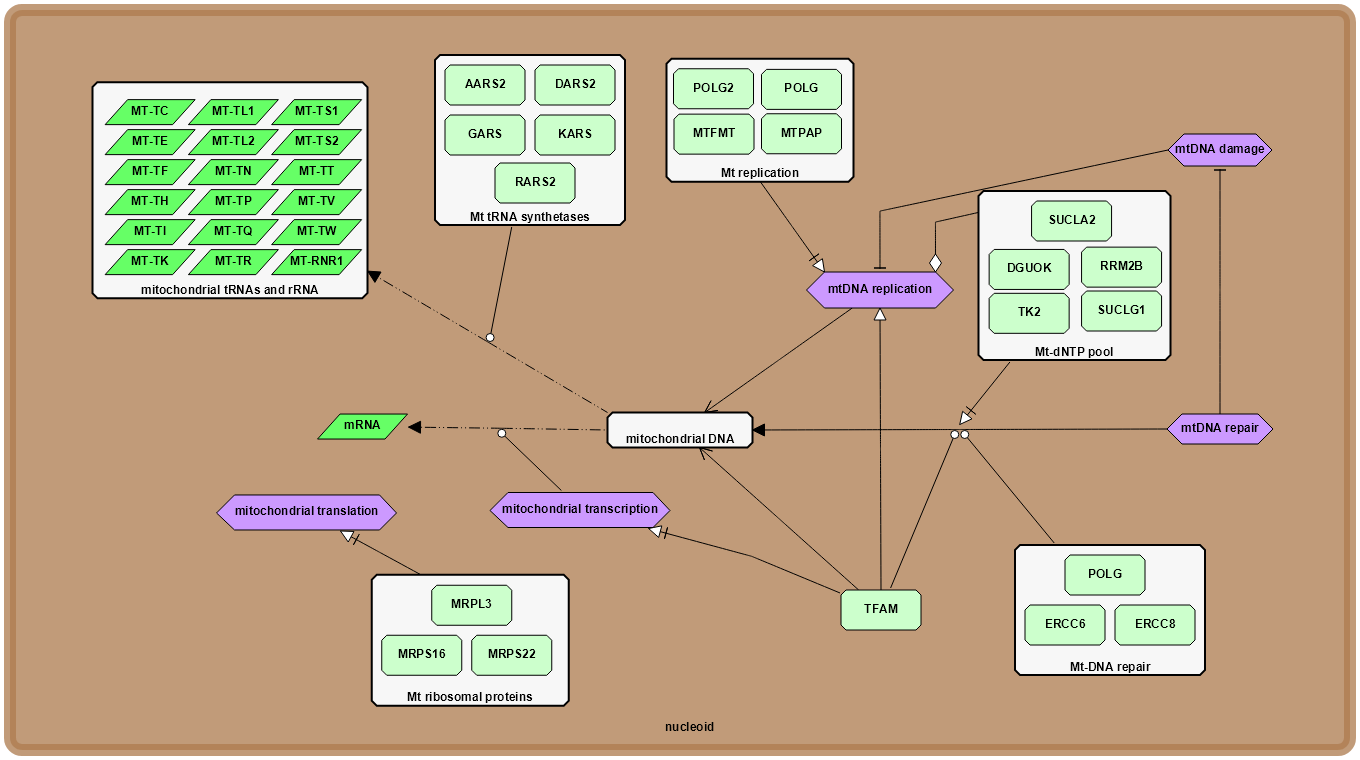 |
Xu, J., Jiang, J., Sauro, H. M.*.
SBMLDiagrams: A Python Package to Process and Visualize SBML Layout and Render.
In: Bioinformatics, 2023.
(JCR Q1, Impact Factor: 4.4)
[Paper] [Code]
|
 |
Welsh, C., Xu, J., et. al..
libRoadRunner 2.0: A High-Performance SBML Simulation and Analysis Library.
In: Bioinformatics, 2023.
(JCR Q1, Impact Factor: 4.4)
[Paper] [Code]
|
 |
Xu, J., Jo J.*.
Broad Cross-reactivity of the T-cell Repertoire Achieves Specific and Sufficiently Rapid Target Searching.
In: Journal of Theoretical Biology, 2019.
(JCR Q2, Impact Factor: 1.9)
[Paper] [Preprint] [Code]
|
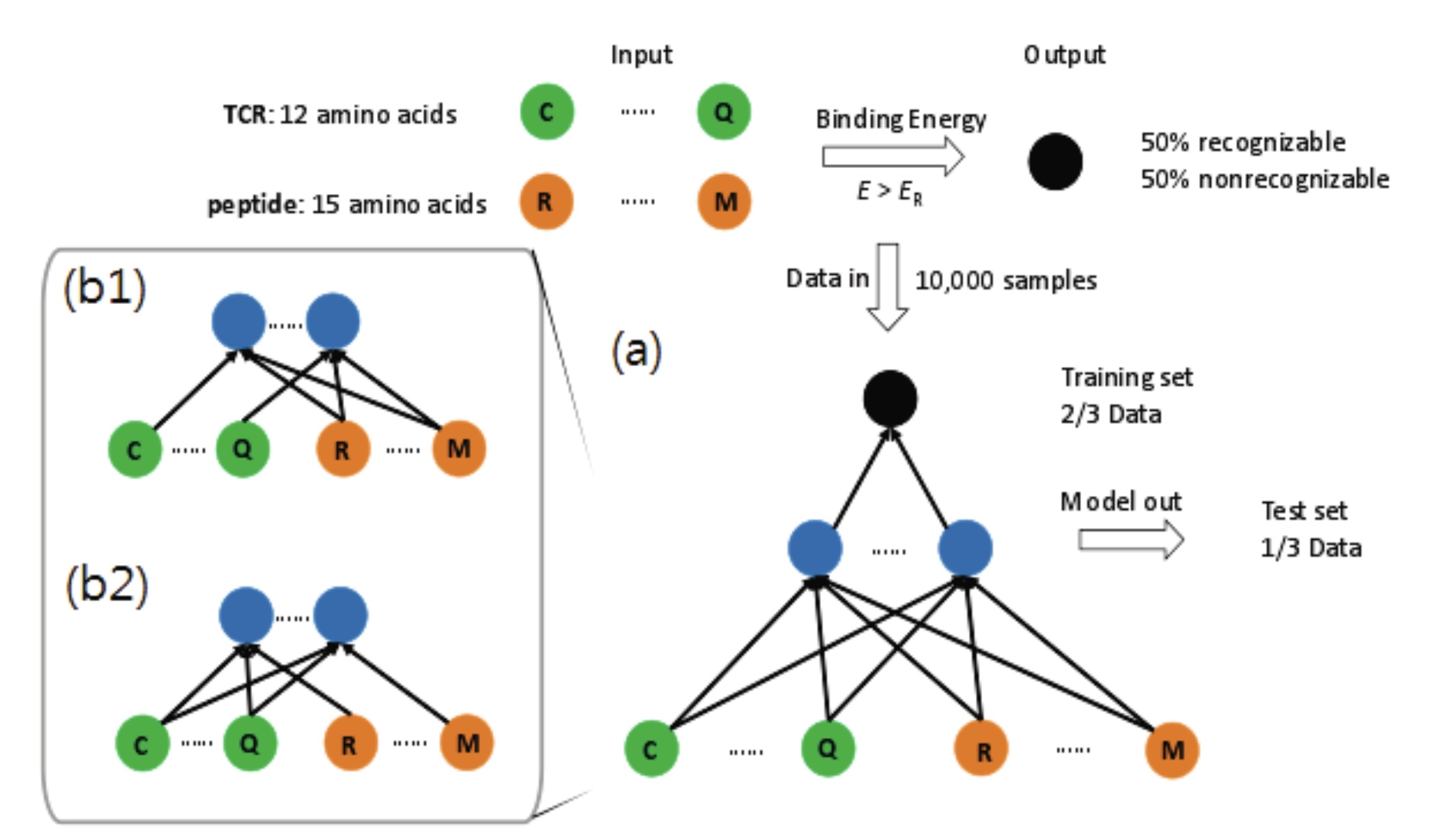 |
Xu, J., Jo J.*.
Immunological Recognition by Artificial Neural Networks.
In: Journal of the Korean Physics Society, 2019.
(JCR Q3, Impact Factor: 0.8)
[Paper] [Preprint] [Code]
|
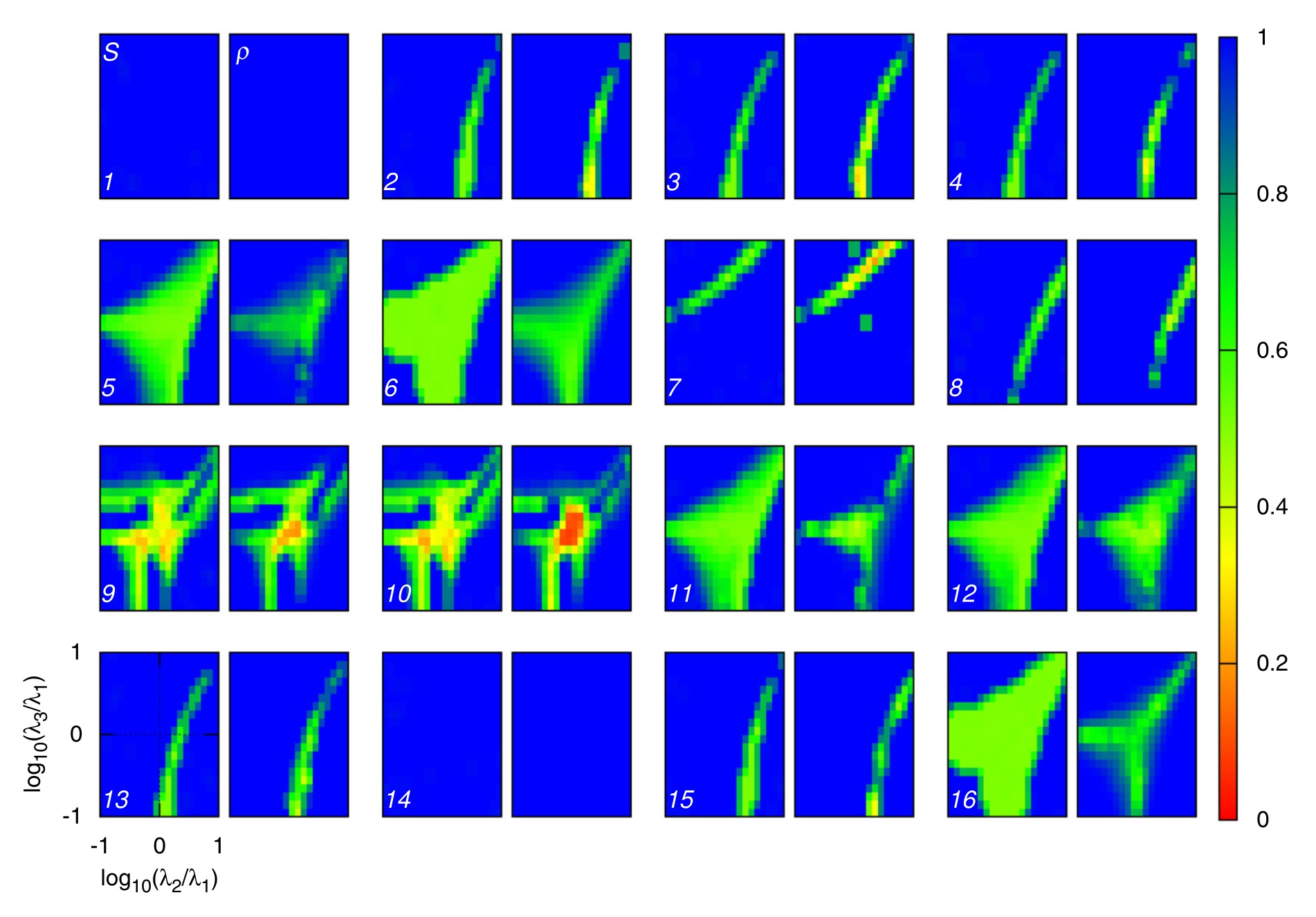 |
Xu, J.#, Park, D. H.#, Jo, J.*.
Local Complexity Predicts Global Synchronization of Hierarchically Networked Oscillators.
In: Chaos, 2017.
(JCR Q1, Impact Factor: 2.7) [# Equal contribution]
[Paper] [Preprint]
|
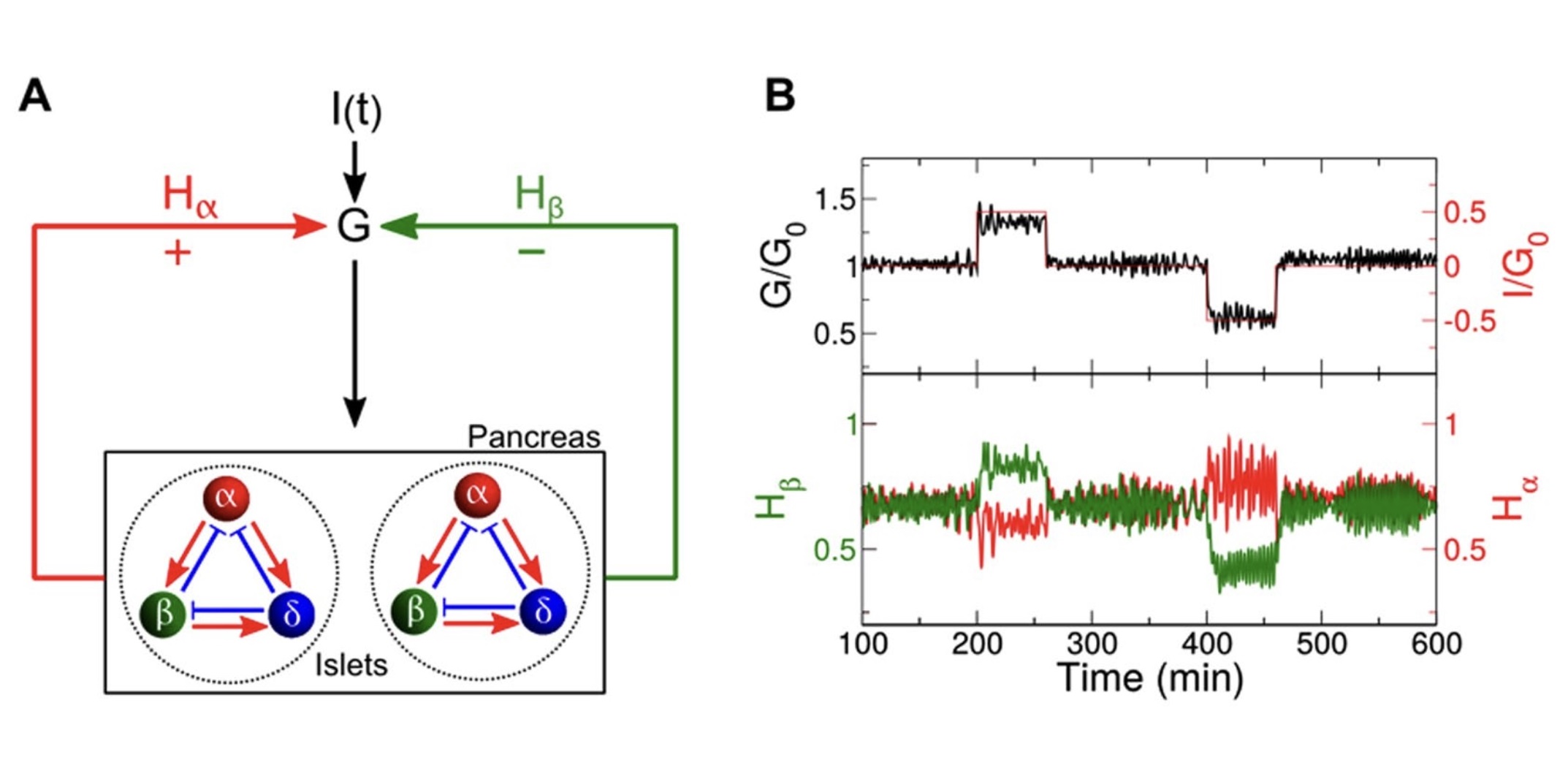 |
Park, D. H.#, Song, T.#, Hoang, D. T.#, Xu, J., Jo, J.*.
A Local Counter-Regulatory Motif Modulates the Global Phase of Hormonal Oscillations.
In: Scientific Reports, 2019.
(JCR Q1, Impact Factor: 3.8) [# Equal contribution]
[Paper]
|
Services
Reviewer for Applied Physics Reviews (IF: 11.9, JCR Q1)
Reviewer for Frontiers in Cell and Developmental Biology (IF: 4.6, JCR Q1)
Reviewer for Biochemical Society Transactions (IF: 3.8, JCR Q2)
Reviewer for BMC Bioinformatics (IF: 2.9, JCR Q1)
Reviewer for Frontiers in Neuroinformatics (IF: 2.5, JCR Q2)
Reviewer for Biosystems (IF: 2.0, JCR Q2)
Reviewer for Journal of Open Source Software









